High performance computing in chemistry
ReaDivivus is a massively parallel software system designed to run on
multicore CPU-s, one or more GPU-s and computing clusters.
It can numerically solve large scale deterministic and stochastic
reaction-diffusion problems up to 3D.
The problem - Reactions, diffusion and patterns in nature
- How can our genes determine the shape of our face or hand (the problem of morphogenesis)?
- How are the different patterns of animal coatings formed?
- What determines the pigment patterns formed on the wigs of butterflies?
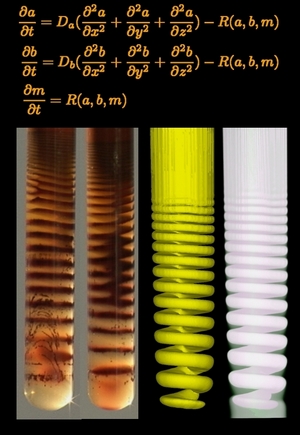
- Where do the stripes of agate rocks come from?
- What about Liesegang rings?
The answer for these and similar questions is usually a properly constructed reaction-diffusion mechanism that mathematically can be formulated as a set of coupled non-linear partial differential equations. Unfortunately these equations usually cannot be treated by standard analytical mathematics. Due to this mathemaical complexity there are many unsolved problems on this field of science.
The promise - Mathematical description
Construction of a possible RD-mechanism and the corresponding equations is a relatively easy task. However due to the above mentioned mathematical problems its validity can be checked only by computer simulation.
RD-systems are generally far too complex for human thinking. One cannot easily predict the possible effects that such a system can produce. At the same time when we try to explain a certain RD problem there will be many possible models that are based on chemically or physically completely plausible effects and have only one drawback: they simply do not work. In these cases the only thing we can do is to construct a slightly different model, and try it again and again.
The solution - Numerical simulation and HPC
ReaDiVivus is a toolset that can help this trial-and-error process, because in most of the cases formulation of a new mechanism requires only several lines of new code.
This software system has grown out from my PhD work and a subsequent scientific project. The original goal was the elucidation of the mechanism of periodic precipitation, often called Liesegang phenomenon.
It is important to emphasize that the results presented on the above link were obtained using the original sequential version of the ReaDiVivus system, which also included some parallel services, but these were based on the PVM system, as general purpose GPUs did not even exist at that time. (PVM stands for Parallel Virtual Machine. It was developed by Oak Ridge National Laboratory and was often called "The poor man's supercomputer" at that times.)
The world has come a long way since then, so of course the ReaDiVivus system can no longer be compared to what it was in 2003. Some operations can now be performed thousands of times faster.
Simulation of the 3D Liesegang spiral shown on the right took 4 days on a
4 CPU cluster.
Today with GPU-s it takes about the same number of minutes.
GPU computing in chemistry - Short list of features
- Simulations can be performed in 1, 2 and 3 dimensions.
- Simulation medium of arbitrary shape can be used.
- Both multicore (CPU) and manycore (GPU) processing is supported.
- All computationally expensive operations are GPU-enabled and can combine the computing power of multiple GPUs
- GPU-related procedures are available in both CUDA and OpenCL, so the system supports both NVIDIA and ATI hardware.
- Simulation workload can be distributed among multiple GPU-s.
- The whole framework uses standard parallelization techniques therefore can be adapted to any supercomputing (HPC) environment.
- The details of GPU-computing are hidden behind wrapper functions so as the user does not have to deal with it (or even know about it).
- During compilation the framework automatically optimizes itself for the actual computing environment.